The Cloning Series: Gibson Assembly
- Subham
- Jul 16, 2020
- 2 min read
#GibsonAssembly was formulated by Daniel G. Gibson back in 2004 when he tried to assemble the Mycoplasma genitalium #genome. The first set of #enzymes were modified later on but the use of three different type of enzymes together to #clone several hundred bps is what made it no less than a #feat back in the day!
Principle
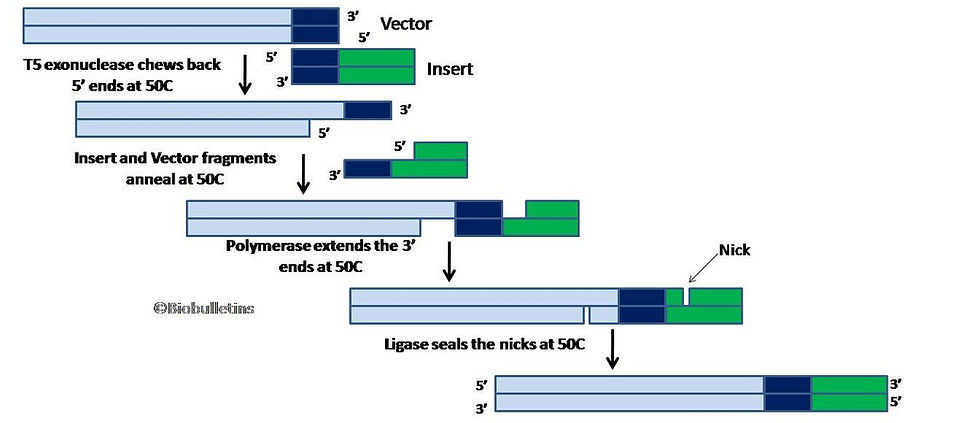
The #GibsonAssembly reaction mix consists three different enzymes:
an exonuclease (#T5exonuclease), which nibbles back the 5’ ends of the DNA fragments (of both insert and vector), generating long #ssDNA overhangs with homology to anneal together
a polymerase (#PhusionPolymerase), to fill in the gaps on the annealed area
a DNA ligase (#TaqLigase), to seal the nicks of the annealed and filled-in gaps, producing single, continuous circular #DNA.
At 50C, the mix does the following:
Protocol
A. Insert PCR
The primer designing for #InsertPCR and #VectorPCR is very crucial. There are certain considerations here like:
a. length should be 18-25 nucleotides keeping in mind 16-80 bp overlapping #homologous regions and #GCcontent must be 40-60%.
b. 3' end of primers for insert PCR should be gene-specific and for vector PCR must be vector specific.
c. Tm should be between 58-65C and difference of Tm between Forward and Reverse primers must be <=4C.
d. The last 5 nucleotides at the 3' ends should contain no more than two G/C.
After PCR, #GelExtraction of the fragment and quantification should be done.
B. Vector PCR
Vector PCR products should be long migrated in the gel to identify the correct linear band to excise and gel extract. After gel extraction it is recommended to do a #DpnI digestion step at 37C to get rid of any circular #plasmid.
C. Gibson Assembly Reaction
This is simple, just add calculated amount of insert and vector, #GibsonAssemblyMix (Miller Lab has a DIY protocol) and water (if needed) to make up a reaction volume of 10-20uL. The reaction is recommended to be incubated at 50C for ~1 hour and must immediately be placed on ice.
D. Transformation
Transform 2.5uL of the reaction into 50uL of highly competent E. coli cells of your choice. Follow your standard #transformation protocol, plate the cells with appropriate antibiotic and incubate at 37C overnight.
E. Colony PCR/ Sequencing
Among the colonies >95% are expected to be transformants. This can be validated through colony PCR with one vector-specific and one insert-specific primer. Colony sequencing may also be done with those sets of primers.
Merits
No requirement of specific #restrictionendonuclease sites
Possible with multiple DNA fragments, although >5 fragments often meet failure!
It enables #DirectionalCloning
Demerits
It is not possible with DNA insert fragments <200bp, possibly because of the chewability of #T5exonuclease!
It requires meticulous primer design
Hard to troubleshoot, always keep a backup plan ready!
In case of multiple fragments, you cannot interchange fragment positions because of the #unique overlapping regions!
Applications
Assembly of large #DNA fragments
Assembly of chemically synthesized single stranded #oligonucleotides into #dsDNA.
More to chew:

Comments