#MeltCurve analysis is done in #qPCR when DNA intercalating dyes like #SYBRGreen, #EvaGreen etc. are used because it will detect any #dsDNA (#primerdimers, Non-specific PCR products, contaminating DNA etc.). No, you'll not have to run the qPCR products in agarose gel every time to rule out contamination or primer dimer, following secondary melt curve peaks!
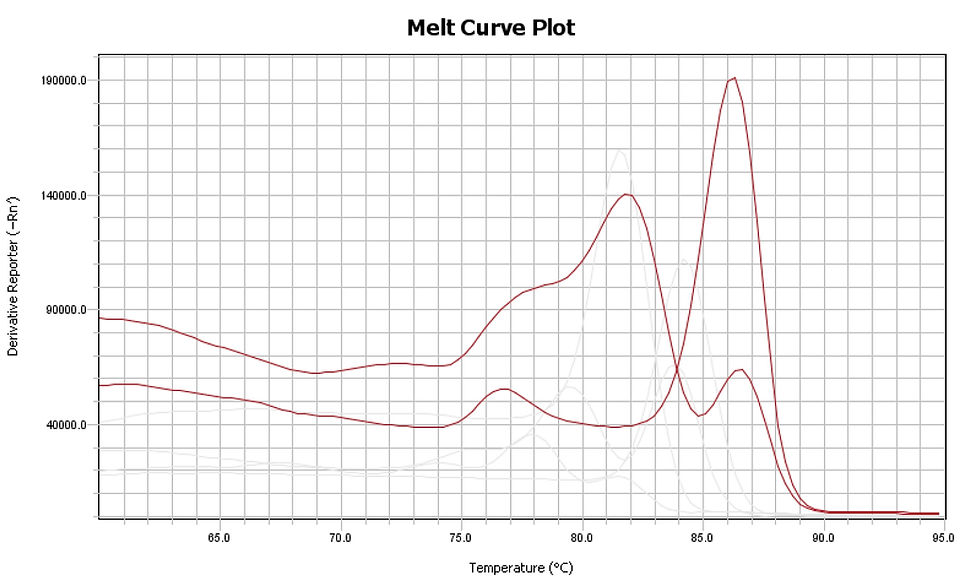
FIGURE: Melt curve plot of two different amplifications with same but varying amount of template. Notice that both has humps/ secondary peaks, but the one with less amount of template DNA has a bigger hump.
What goes wrong?
1. Use of excess primers in DNA intercalating dye based #qPCR almost always fetch you secondary melt curve peaks because of #primerdimers.
2. Presence of contaminating DNA sequences in RNA samples while performing #RTqPCR.
3.There could be #splicevariants of your target genes, having some extra bases in your target region, getting you a higher melting temperature for your PCR product, and consequently a secondary #meltcurvepeak.
Solutions?
a. Use appropriate amount of primers, to standardize your assay you can perform one #primergradient amplification with every other component unchanged. Doing a primer gradient assay from 0.25uM to 2uM with 0.25uM graduation could be useful to start with. You may find other useful suggestions regarding primer dimers here.
b. Treat your RNA with DNase before proceeding to RTqPCR, this will help to keep contaminating genomic DNA in check, or set up a -RT control.
c. For contaminated RTqPCR, you can also change your contaminated PCR Mastermix and primer stocks, and use #barriertips for avoiding contaminating genomic DNA.
d. Designing primers specific to splice variants will ensure you do not get a larger or shorter amplicon, and thus lesser probability of #doublepeaks.
All done and said, #secondarymeltcurvepeaks can also arise from a #triphasic melting of DNA i.e. apart from the double-stranded and single-stranded forms there could be an intermediate form where the A-T bonds have been broken but G-C bonds are still intact. This happens when the amplicon has AT-rich subdomains causing #unevenmelting. In that case, you can use the uMelt tool, which is computational predictive tool to find amplicon melting nature.
Hope it helps!




Comments